These musings are written by a computer scientist who has recently been studying for an MSc in biochemistry / molecular biology, and came across some interesting features of human mitochondrial DNA, which are comparable with clever software engineering (also known as a "cool hack").
Humans have two repositories for genes (instructions coded in DNA) within a cell: chromosomal DNA and mitochondrial DNA. Genes stored in your chromosomes (22 of them, and XX if you are female or XY if male) are linear sequences of nucleotides about 3,000 Mb (million bases) in size. Mitochondrial DNA is a very small circular piece of DNA (about 16kb in size). mtDNA is 16,568 base pairs in length and encodes 13 proteins, 22 distinct transfer RNAs (tRNA) and two ribosomal RNAs (rRNA).
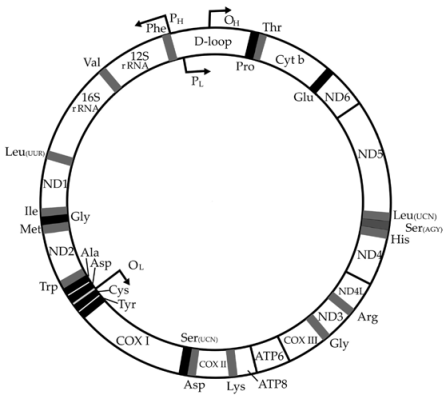
Genes are similar to opcodes or machine instructions used by microprocessors in software to execute programs. It turns out that there are some very cute "hacks" in the mitochondrial DNA, even better than hacks in software. Genes stored as DNA code for both RNA and polypeptide chains of amino acids, which fold to form proteins. The cute hack is that the coding for two proteins, ATP synthase6 and ATPase8 (shown right at the bottom of the circular mitochondrial map) are encoded by the same nucleotides, but using different offsets in the mitochondrial DNA.
In software, usually, 8 bits = 1 bytes, and depending on the processor, say, 32 bits = 1 opcode. If you started reading the opcode in the middle of those bits, you'd execute a completely different instruction, e.g. execute the 6502 instruction BNE (branch not equal) instead of a STA (store accumulator)
This is exactly what happens when coding for ATPase6 and 8. DNA codes for mRNA in base 4 (G, C, T, A) rather than binary (0 or 1), and each codon ("opcode") is 3 bases long to code for an amino acid. Only they overlap but use different "reading frames" -- much like two executable images sharing the same byte stream, but running different opcodes, because the offsets were different. Like encoding NOP, LDA, BNE for one executable, and PLA, PLY, JSR using the same bits but read from a different offset.
This is very, very cute and clever. And they both produce fully functional proteins (i.e. working code!). I would say your mitochondrial DNA is our "bootstrap prom" code, since it's very small, and only encodes a few essential proteins and RNAs, but is very compact and tightly coded. Your chromosomal DNA is much more like a Microsoft Word executable -- bloated, millions of bases/bytes long, written in C++ and hence lots of "junk" and padding in the executable, and can tolerate a few corrupted bytes (nonsense mutations, frameshift errors, etc) here and there.
See also Lucy McWilliam's Why DNA is better than Perl page.